A small molecule is likely to interact with protein in a concave region of the surface of the protein. There are various ways to identify concave regions on protein surface. This website employs a modified Cx algorithm, the simplest way to identify concaves.
The original Cx algorithm introduced by Pintar et al (2002) is to identify protruding atoms in proteins. The principle of the algorithm is illustrated in Figure 1. For each heavy atom in a protein structure, the number of heavy atoms within a fixed distance R is calculated, the number is multiplied by the mean atomic volume found in general proteins(20.1±0.9Å3 ;Richards,1974), and obtain the volume Vint occupied by protein atoms within the sphere. The remaining volume of the sphere (Vext) is calculated as the difference between the volume of the sphere and Vint. Pintar et al defined Cx value as;
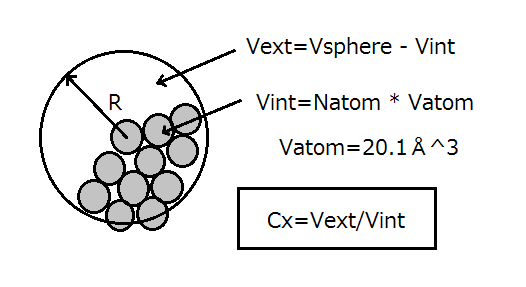
Fig.1 Cx algorithm
To identify a concave region, Cx is be calculated as above with the radius
of 12~20Å and the region with low Cx value is taken as a candidate.
We further modified the original method in the following two points;
- We modified the volume of sphere by adding 2.0Å to the radius, because heavy atoms on the surface of the sphere partially penetrate out of the sphere hence the sum of the atomic volume exceeds the volume of the sphere when the atoms are fully packed.
- We added 0.19 to any Cx values. When we calculated the value of Cx on artificially built polycarbon in a computer with 2.0Å interatomic distances, the minimal value of Cx was about -0.19 (Figure 2). This negative Cx value is considerd to be caused by overlapping of the carbon sphere of a van der Waals radius, so that for practical purposes we added 0.19 to any Cx values.

Fig.2 Artificially designed polycarbon
(2.0Å interatomic distance)